What we do?
Morphogenesis is the process that generates the shape of tissues and organs. Our lab seeks to elucidate the molecular and cellular basis of epithelial tissue morphogenesis to better understand basic mechanisms of animal development.
Our Mission
Motivation
Morphogenesis is controlled primarily by local coordinated changes of cell shape, rearrangements of cell-cell contacts, cell proliferation and cell death. The genes and molecular mechanisms that regulate these cellular behaviors are only partially characterized. Therefore, a major goal is to identify novel genes that participate in tissue morphogenesis and elucidate their mechanism of function. Anomalies in epithelial morphogenesis underlie a range of congenital diseases such as spina bifida, cystic kidneys and vascular aneurysm, and acquired diseases such as cancer that disrupt the normal morphology of tissues and organs. To understand these disease processes a deep understanding of the mechanisms involved is essential.
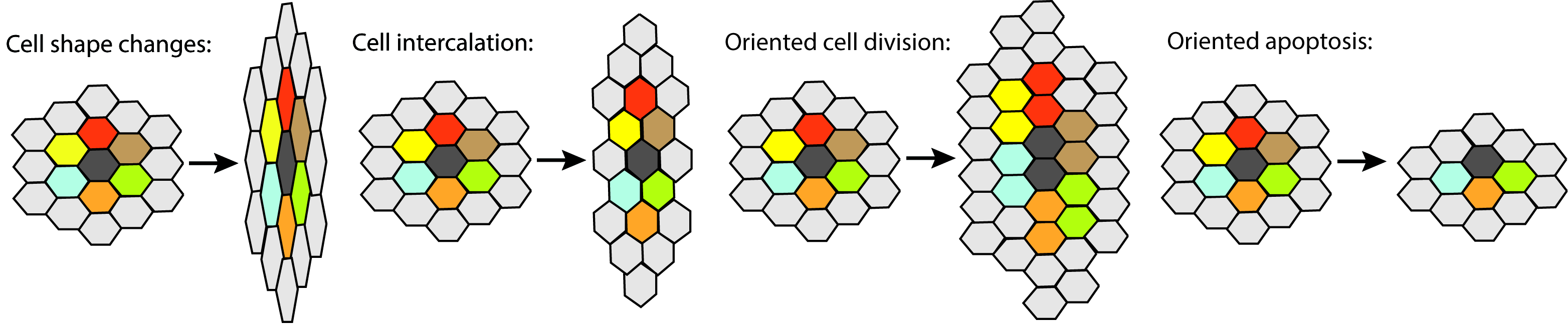
Coordinated cell behaviors drive tissue morphogenesis. Coordinated changes in cell shape, rearrangements of cell-cell contacts, cell proliferation and cell death lead to global changes in tissue morphology. Our lab employs genetic analysis, live imaging and quantitative image analysis to elucidate the molecular and cellular basis of epithelial morphogenesis.
Coordinated cell behaviors drive tissue morphogenesis
Critical barriers to knowledge
Mechanical forces generated by cell proliferation, apoptosis, cell-cell adhesion and the cytoskeleton drive morphogenesis. However, the modulation of mechanical force generation in space and time by extracellular signals, polarized membrane landmarks and cytoskeletal effectors are just being elucidated. The convergence of recent advances in imaging technologies, fluorescent reporters, computational image analysis and mathematical modeling enables developmental biologists to observe and define the molecular and cellular basis of morphogenetic processes with unprecedented detail.